Xinyu Huang1, Kimiaki Shirahama2, Frederic Li1, Marcin Grzegorzek1


1 Institute of Medical Informatics, University of Luebeck, Germany
2 Department of Informatics, Kindai University, Japan
Note (last updated on April 18, 2023): This webpage is a copy of the one linked in the paper, which was originally located on the website of Kindai University (Japan). Because of the movement of the second author who is managing this webpage, the original webpage will no longer be accessible shortly. Consequently, all research materials used in the paper have been moved to the current institution of the second author Doshisha University (Japan). The contents contained and linked in this webpage are exactly the same as the original ones.
The studies show that the prevalence of sleep disorder in children is far higher than that in adults. Although much research effort has been made on sleep stage classification for adults, children have significantly different characteristics of sleep stages. Therefore, there is an urgent need for sleep stage classification particularly targeting children. Our research focuses on two issues: The first issue is timestamp-based segmentation (TSS) to deal with the fine-grained annotation of sleep stage labels for each timestamp. Compared to this, popular sliding window approaches unnecessarily aggregate such labels into coarse-grained ones. We utilize DeConvolutional Neural Network (DCNN) that inversely maps features of a hidden layer back to the input space to predict the sleep stage label at each timestamp. Thus, our TSS-based DCNN can boost the classification performance by considering labels at numerous timestamps. The second issue is the necessity of multiple channels. Different clinical signs, symptoms or other auxiliary examinations could be represented by different Polysomnography (PSG) recordings, so all of them should be analyzed comprehensively. Hence, we exploit multivariate time-series of PSG recordings, including 6 electroencephalograms (EEGs) channels, 2 electrooculograms (EOGs) channels (left and right), 1 electromyogram (chin EMG) channel and double leg's electromyogram channels. Our DCNN-based method is tested on our SDCP dataset collected from child patients aged from 5 to 10 years old. The results show that our method yields the overall classification accuracy of 84.27% and macro F1-score of 72.51% which are higher than those of existing sliding window-based methods. One of the biggest advantages of our DCNN-based method is that it processes raw PSG recordings and internally extracts features useful for accurate sleep stage classification. We examine whether this is applicable for sleep data of adult patients by testing our method on a well-known public dataset Sleep-EDFX. Our method achieves the average overall accuracy of 90.89% that is comparable to those of state-of-the-art methods without using any hand-crafted features. This result indicates the great potential of our method because it can be generally used for timestamp-level classification on multivariate time-series in various medical fields. Additionally, we provide source codes so that researchers can reproduce the results in this paper and extend our method.
This paper offers three main contributions:

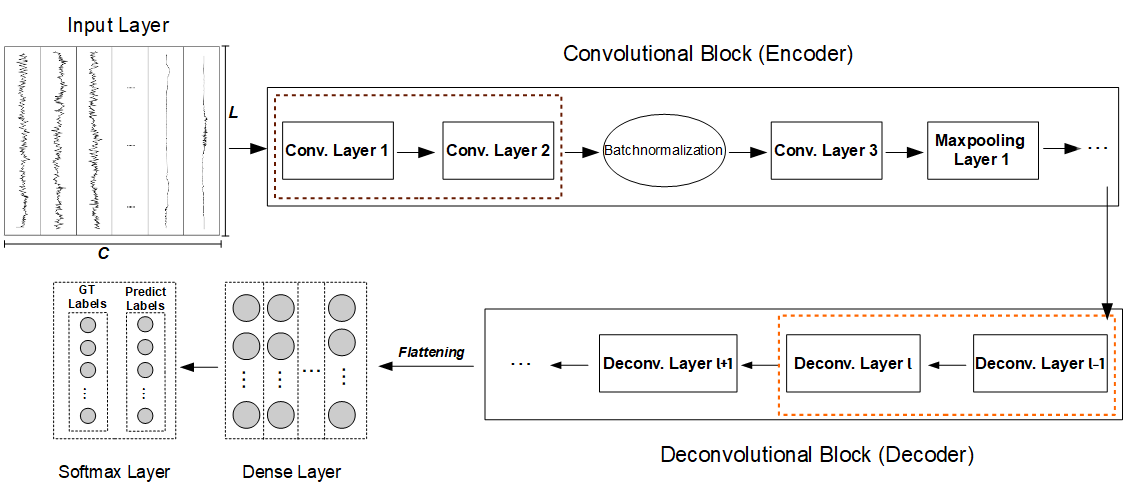
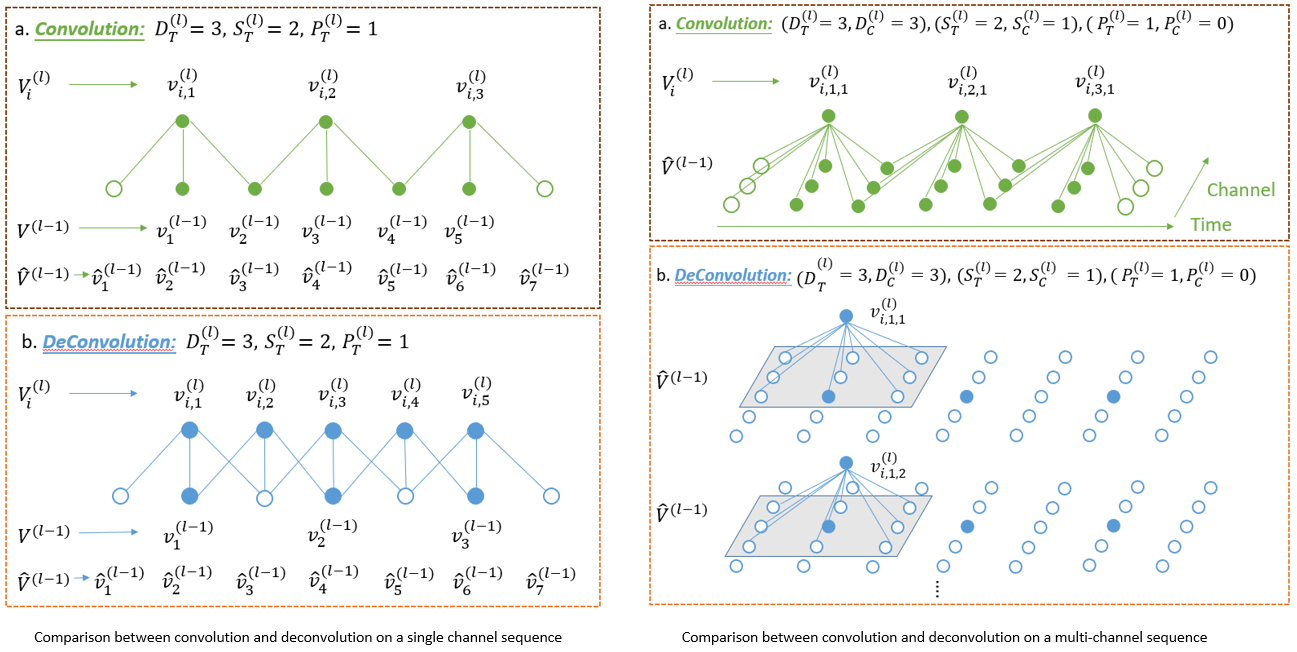
We use a DCNN that refers to a kind of neural network composed of convolutional and deconvolutional blocks that work as an encoder and decoder, respectively. The convolutional block in the encoder achieves a downsampling, which can condense an input sequence, reduces the amount of data size and ignores subtle noise information. Hence, a feature map resulting from the encoder is an abstracted representation of the input sequence. Then, the decoder takes it as input and utilizes deconvolutional layers, each of which translates a coarse feature map into a denser and detailed one while preserving a connectivity pattern that is compatible with the convolution. In other words, the decovonlutional layer extends the dimension of the abstracted feature map by distilling latent features that are ignored through the convolutional block, so that the sleep stage label at each timestamp is approximated. Our DCNN is outperformed than other DNN models.
In our experiments, we use the SDCP dataset that includes 21 child patient subjects aged from 5 to 10 years old (14 females and 7 males). We collected sleep data using a patients monitoring system named Philips Alice 6 made by Loewenstein Medical . This system is very flexible and can easily manage every patient's room. Three types of sensor devices were used for the data collection in our research. The details of these three sensor devices are described below:
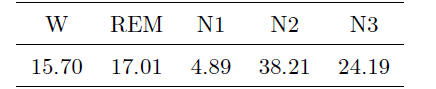
We use four sensor modalities provided by these sensor devices as mentioned in the above listings, including 6 EEG (O1M2, O2M1, F3M2, F4M1, C3M2, C4M1) channels, 2 EOG (left and right) channels, 1 chin EMG channel, and double legs channels. The sampling frequency of all sensors is 200Hz and the length of collected multi-channel sequences is around 10 hours from the evening to the next day morning. Corresponding hypnograms (labels) were manually scored by experienced scorers using the American Academy of Sleep Medicine (AASM) scoring rules. The percentages of labels of each sleep stage in our dataset are shown in the following Table:
We carried out a comparative experiment using our proposed TSS-based DCNN model on the Sleep-EDFX dataset[1,2]. Sleep-EDFX is a well-known public available dataset that contains 197 whole-night sleep PSG recordings, including EEG (Fpz-Cz and Pz-Oz defined by combining orange-colored electrodes in Fig. 1), horizontal EOG, and chin EMG. All data come from 2 studies, e.g., Sleep Cassette Study (SC) and Sleep Telemetry Study (ST). In SC, a total of 153 recordings were collected between 1987 and 1991 to study the effects of age on sleep of healthy Caucasians who are 25 to 101 years old and take no medications. In ST, 44 recordings were obtained in 1994 to study the effects of temazepam medications on sleep in 22 Caucasians. All subjects have mild difficulties falling asleep but were otherwise healthy. The 9 hours PSG recordings were obtained in the hospital during two nights. Subjects took temazepam on one night and a placebo on the other night. The sampling frequency is 100Hz. Corresponding hypnograms (labels) were manually scored by experienced scorers using the Rechschaffen & Kales (R & K) scoring rules.
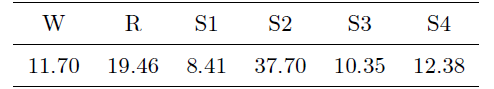
In our experiment, we decide to randomly select 12 PSG recordings of 6 subjects in the ST study, including 2 EEG (Fpz-Cz and Pz-Oz) channels and a horizontal EOG channel to implement a comparative experiment. We consider four classification cases, 6-stage classification targeting W, S1, S2, S3, S4 and R stages, 5-stage classification where S3 and S4 in 6-stage classification are combined into one stage, 4-stage classification where S1 and S2 in 5-stage classification are merged, and 3-stage classification just considering W, NREM (S1, S2, S3, S4) and R stages. The proportion of sleep stages in our selected Sleep-EDFX dataset is shown in the following Table:
For management purposes, if you want to download the dataset and codes, please contact the corresponding author of the publication via email (huang@imi.uni-luebeck.de) first.
For any question about the paper, study or codes, please contact the corresponding author of the publication.